One of the world’s most lethal families of plant viruses has been revealed in unprecedented detail in a new study that may provide clues to preventing the global spread of the pathogen.
The complex 3D structure of the geminivirus is revealed in the joint study carried out by researchers at the University of Leeds and the John Innes Centre.
Geminiviruses are responsible for diseases affecting crops such as cassava and maize in Africa, cotton in the Indian subcontinent and tomatoes across Europe.
Being able to see its structrure in great detail is vital as it could help virologists and molecular biologists better understand the virus lifecyle, and develop new ways to stop the spread of these viruses and the diseases they cause.
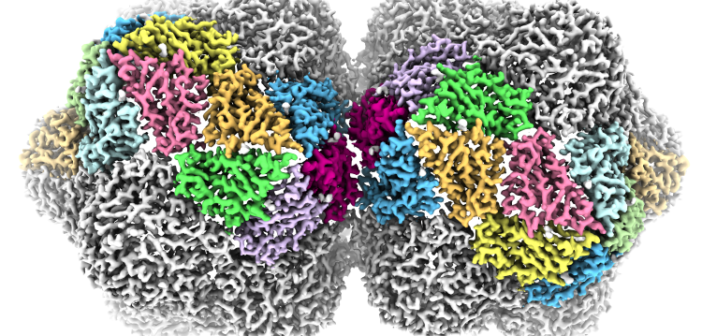
These viruses are named for their curious shape. Viruses usually have a protective shell of protein, or capsid, that acts to protect their genetic material in the environment. In most viruses, this capsid is roughly spherical, but the geminivirus has a ‘twinned’ capsid formed by two roughly spherical shapes fused together.
The molecular details of how this twinned capsid is achieved – and how it assembles in cells or expands to release the genome and start a new infection – has remained a mystery, despite the risk posed by the virus to agricultural economies worldwide.
Researchers at the Leeds University’s Astbury Centre for Structural Molecular Biology used cryo electron microscopy techniques to study geminivirus structure at undprecedented resolution, and in the process have begun to untangle its assembly mechanisms.
Published in Nature Communications, the study reveals how the capsid of the geminivirus is built and how its single-stranded DNA genome is packaged.
“In many other types of virus, the spherical capsids are built from a single protein that adopts three different shapes, which then fit together to form a closed container,” explains Professor Neil Ranson, who led the research team at the Astbury Centre.
“But geminivirses are not spherical, so must be using a different set of rules. Using cryo-EM, we’ve been able to show that they do use three different shapes of the same protein, but with a completely different rulebook for assembly”
One of the difficulties in studying geminviruses is growing them in sufficient quantities for structural studies.
The team studied a type of geminvirus transmitted by whitefly called ageratum yellow vein virus, which was produced in tobacco plants under carefully controlled conditions by researchers at the John Innes Centre.
The team at the John Innes Centre led by Dr Keith Saunders and Professor George Lomonossoff also developed a method for assembling geminivirus particles within plants in the absence of infection.
This highlighted the role played by the single-stranded DNA in particle formation.
“Having worked for many years to understand the diseases geminiviruses cause, it was very satisfying to apply modern genetic methods to generate these geminate structures,” said Dr Saunders.
“The big surprise arising from this study was that fact that the virus coat protein can adopt different conformations that are dependent upon its location in the structure – it is different at the equator than at its apexes. That helps to explain how the particles form during virus infection. With this new knowledge, it now means that future studies can be directed to seek ways to disrupt geminate structure maturation by making antivirals that target those areas.”
Dr Suanders said the John Innes Centre team had been studying ageratum yellow vein virus for 20 years and their knowledge of the diease made it a good candidate for closer inspection.
It is part of white-fly transmitted group responsible for tomato yellow leaf curl disease, a disease affecting tomato production in many countries around the Mediterranean Sea and cotton leaf curl disease affecting cotton plants in India and Pakistan.
Both diseases give rise to tremendous crop losses and so are economically very damaging.”
The work was funded by the Biotechnology and Biological Sciences Research Council.